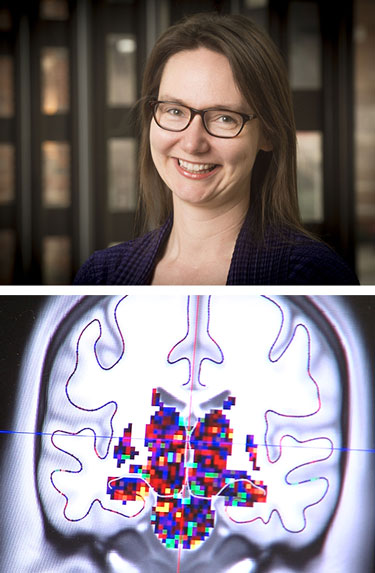
 Kristina Visscher (above) is using fMRI images from several hundred brains to learn how the brain adapts after long-term changes in vision.What do the human brain, the 3 billion base-pair human genome and a tiny cube of 216 atoms have in common?
Kristina Visscher (above) is using fMRI images from several hundred brains to learn how the brain adapts after long-term changes in vision.What do the human brain, the 3 billion base-pair human genome and a tiny cube of 216 atoms have in common?
All of them, from the tiny cube to the 3-pound human brain, create incredibly complex computing challenges for University of Alabama at Birmingham researchers, and aggressive investments in UAB’s IT infrastructure have opened new possibilities in innovation, discovery and patient care.
For example, Kristina Visscher, Ph.D., assistant professor of neurobiology, UAB School of Medicine, uses fMRI images from several hundred human brains to learn how the brain adapts after long-term changes in visual input, such as macular degeneration. Frank Skidmore, M.D., an assistant professor of neurology in the School of Medicine, studies hundreds of brain MRI images to see if they can predict Parkinson’s disease.
David Crossman, Ph.D., bioinformatics director in UAB’s Heflin Center for Genomic Science, deciphers the sequences of human genomes for patients seeking a diagnosis in UAB’s Undiagnosed Diseases Program, and he processes DNA sequencing for UAB researchers who need last-minute data for their research grant applications.
Ryoichi Kawai, Ph.D., an associate professor of physics in the UAB College of Arts and Sciences, is laying the groundwork for a better infrared laser by calculating the electronic structure for a cube made up of just 216 atoms of zinc sulfide doped with chromium or iron.
Each researcher faces a mountain range of computational challenges. Those mountains are now easier to scale with UAB’s new supercomputer — the most advanced in Alabama for speed and memory.
The tiny cube
The 216-atom electronic structure problem “is the largest calculation on campus, by far,” Kawai said. “If you give me 2,000 cores, I can use them. If you give me 10,000 cores, I can use them all without losing efficiency.” (UAB's new supercomputer has 2,304 cores.)

|
Brains
In their research, both Skidmore and Visscher have to compare brains with other brains. Because each brain differs somewhat in size, shape and surface folds, every brain has to be mapped onto a template to allow comparisons.
Physicist Kawai describes his computations as the most intensive on campus.“We want to capture information in an image, such as information on an individual’s brain condition,” Skidmore said. “The information we are trying capture, however, can often be difficult to see in the sea of data we collect. One brain may contain millions of bits of data in the form of ‘voxels,’ which are a bit like the pixels on your TV but in three dimensions.”
“When we map to a template,” Skidmore said, “we quadruple the data. When we ask, ‘How does this compare to a healthy brain?’ we double the data again. Then if we look across one brain or across multiple kinds of brain images, the amount of data truly explodes.”
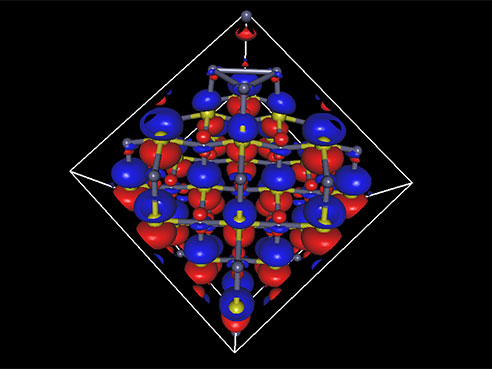 Complex calculations of the electronic structure of a cube of atoms are laying the groundwork for better lasers.“This is made even more complex by the fact that a given image can include more than three dimensions of information,” he said. “One type of image we use generates 5-dimensional brain maps. Since we can’t see in five dimensions, we ask the computer to work in these higher-dimensional spaces to help us pull the information out of the data.”
Complex calculations of the electronic structure of a cube of atoms are laying the groundwork for better lasers.“This is made even more complex by the fact that a given image can include more than three dimensions of information,” he said. “One type of image we use generates 5-dimensional brain maps. Since we can’t see in five dimensions, we ask the computer to work in these higher-dimensional spaces to help us pull the information out of the data.”
To look at the adult brain’s plasticity — the ability to change function and structure through new synaptic connections — Visscher studies visual processing.
“Because we look at spatial and temporal data, the number of pieces of information is huge — gigabytes per subject,” she said. “We need to do correlations on all the data points at the same time. To get faster, we optimize the data analysis with a lot of feedback. Then we run what we learned from one brain on a hundred brains.”
UAB customers
Crossman deciphers the sequences of human genomes for patients seeking a diagnosis, and he processes DNA sequencing for UAB researchers. Providing excellent customer service to his clients is vital, Crossman says, and it takes computer power to crunch genome sequencing data for those researchers, physicians and patients.
Managing traffic on the new UAB computer clusterSize:
Queues:
Scheduling
|
“And I cannot tell a researcher, ‘I’m sorry, we won’t meet that grant deadline,’” Crossman said. “You know, science can’t stop.”
The data floodgates of genomics burst open about a dozen years ago with the arrival of next-generation, high-throughput sequencing, says Elliot Lefkowitz, Ph.D., director of Informatics for the UAB Center for Clinical and Translational Science. Lefkowitz has been serving the bioinformatics needs of the UAB Center for AIDS Research for 25 years, and now also handles bioinformatics for the UAB Microbiome Facility. His team has grown to five bioinformaticians and several programmers.
“We deal with billions of sequences when we do a run through the DNA sequencing machine,” Lefkowitz said. “We need to compare every one of the billion ‘reads’ (the 50- to 300-base sequence of a short piece of DNA) to every other one. With high-performance computing and thousands of nodes, each one does part of the job.”
“In not too many years,” Lefkowitz speculated, “we will be sequencing every patient coming into University Hospital.”
Changes like that mean ever-increasing computer demands.
“Biomedical research,” Crossman said, “now is big data.”