Title:
Association of the cervicovaginal microbiome with cervical shortening in women with prior spontaneous preterm birth
Authors:
Akila Subramaniam, MD, MPH, Boyi Guo, MS, Elena Lobashevsky, MD, PhD, William van der Pol, MS, Elliot Lefkowitz, PhD, Casey Morrow, PhD, Jeff M Szychowski, PhD, Nengjun Yi, PhD, John Owen, MD, MSPH.
Background:
- Preterm birth (PTB) is a leading cause of neonatal morbidity and mortality worldwide
- Midtrimester cervical length (CL) shortening is a powerful predictor of and precursor to spontaneous preterm birth (SPTB), especially in women with prior SPTB (high-risk).
- A prevailing hypothesis is that pathologic microbial stimuli from the lower genital tract lead to localized inflammation and subsequent SPTB.
- Culture-independent techniques using 16S rDNA gene sequencing can be utilized to more precisely describe the cervicovaginal microbiome and its relationship to CL shortening and SPTB.
Objective:
- To compare the cervicovaginal (CV) microbiome in high-risk women with and without CL shortening.
Study Design:
- We performed a nested case-control study using stored CV biospecimens previously collected in a multicenter randomized trial of ultrasound-indicated cerclage in high-risk women undergoing serial CL screening at 160–226 weeks
- Women with CL <25mm were randomized to cerclage versus no cerclage.
- Enrolled women also had cervicovaginal fluid samples collected at the first CL measurement, and if not shortened on the initial exam, again at the time of diagnosed CL shortening.
- Samples from the primary trial were stored at -80C.
- For this study, we used CV specimens from the initial study visit in women who later had CL shortening versus those who did not.
- DNA was extracted with appropriate PCR primers and a V4 amplicon library prepared and then sequenced on the MiSeq platform.
- The QIIME package, using DADA2, generated Amplicon Sequence Variants with a high yield of species-level identifications.
- Differential tests were performed at both community level (alpha diversity and beta diversity) and taxa-level microbiome compositions using nonparametric and parametric tests respectively.
- Negative binomial models, adjusting for covariates (maternal age, race, and BMI), were used in taxa-specific analysis
- False discovery rate was used to adjust for inflated type I error.
Results:
- 391 samples met quality standards and included in analysis
- 121 with CL shortening
- 270 without CL shortening.
- A total of 230 distinct operational taxonomic units at the species level were identified.
- There was no significant difference between the two groups in terms of alpha diversity using three distinct analysis measures (Figure 1a)
- There was no statistically significant differences in beta diversity (PERMANOVA p = 0.07) between the two groups (Figure 1b, no significant clustering).
- While no community-level differences were observed, 29 taxa were significantly associated with CL shortening after covariate adjustment (Top 10 in Table).
Conclusion:
- While multiple taxa-level differences were found in the CV microbiome of high-risk women who go on to have midtrimester CL shortening (versus not), community-level differences were not observed.
- Further studies should evaluate if CV microbial changes occur between initial CL evaluation and time of CL shortening and if the identified taxa are associated with a localized inflammatory response.
Figure 1. Comparison of alpha diversity (a. left panel) and beta diversity (b. right panel – Principal Coordinate Analysis) between women with a prior SPTB who go on to experience CL shortening versus those who do not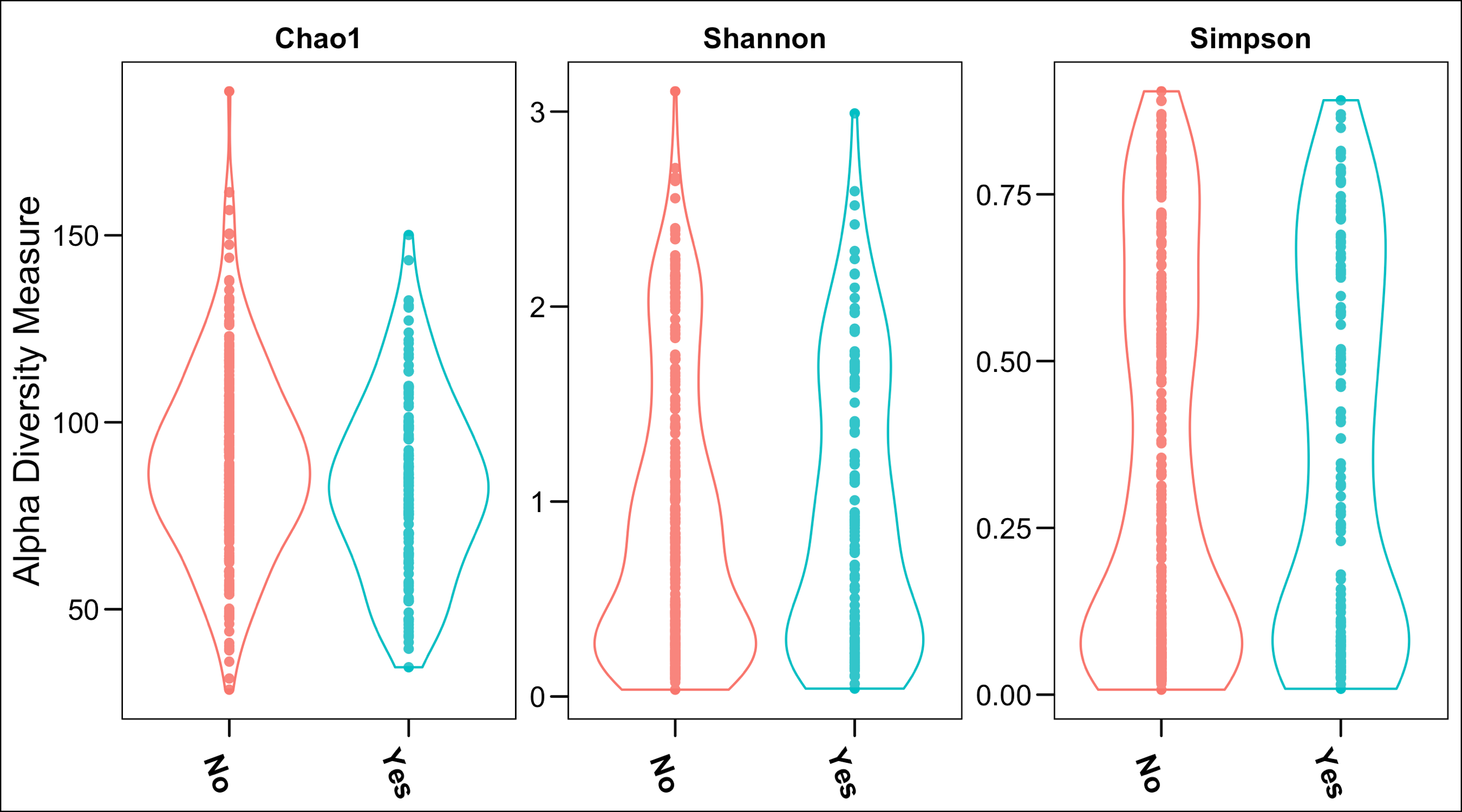
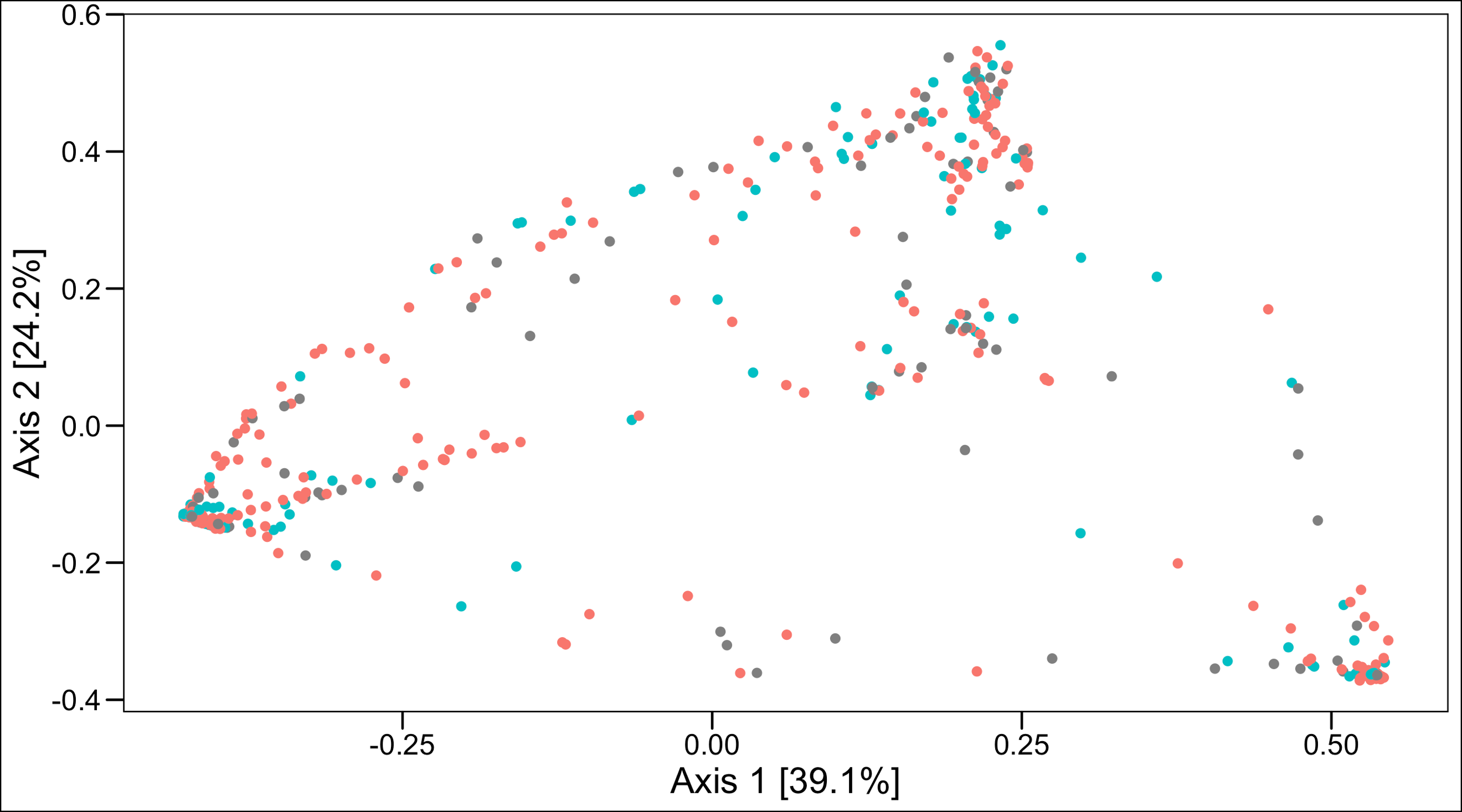
Table 1. Top 10 operational taxonomic units (of 29 statistically significant taxa, FDR p<0.05) with strongest association with CL shortening after covariate adjustment.
Most significant p-value at top
|
Family |
Genus |
Species |
|
Lachnospiraceae |
Blautia |
Un-named |
|
Ruminococcaceae |
Faecalibacterium |
prausnitzii |
|
Bacteroidaceae |
Bacteroides |
Un-named |
|
Lachnospiraceae |
Blautia |
obeum |
|
Lachnospiraceae |
Coprococcus |
Un-named |
|
Ruminococcaceae |
Faecalibacterium |
prausnitzii |
|
Bacteroidaceae |
Bacteroides |
NA |
|
Comamonadaceae |
NA |
NA |
|
Lachnospiraceae |
Roseburia |
NA |
|
Lachnospiraceae |
Blautia |
Un-named |