Media contact: Jeff Hansen
 Jake Chen (left) and James Cimino (right) are aiming to break down scientific data silos with the UAB Biomedical Research Information Technology Enhancement initiative.
Jake Chen (left) and James Cimino (right) are aiming to break down scientific data silos with the UAB Biomedical Research Information Technology Enhancement initiative.
When your research team extends from the Caribbean to Chesapeake Bay and is responsible for hundreds of billions of DNA base-pairs of data, you have more than a scientific mystery on your hands — you have a logistical tangle, as well.
Jeffrey Edberg, Ph.D., professor in the UAB School of Medicine Division of Clinical Immunology and Rheumatology and director of the Center for Clinical and Translational Science (CCTS) Specimen Processing, Analysis, and Biorepository unit, is a member of a long-term clinical study of systemic lupus erythematosus (SLE) that has enrolled more than 8,600 patients and healthy controls at UAB and other institutions from Baltimore to Puerto Rico.
|
"We had an enormous collection of data. The problem was it was all split up." |
The project’s data trove includes more than 8,000 genomewide association studies, more than 6,500 copy number variant assays, 600 targeted exome sequencing studies, more than 75 whole genome sequencing studies and a host of biobanked samples from almost all study participants, including plasma and serum. Finding new connections among all of those information sources could offer important clues to understanding the puzzling variety in lupus cases, including why the disease occurs more frequently in women and why minority patient populations have worse outcomes than other groups.
"We had an enormous collection of data," Edberg said. "The problem was it was all split up." That is, until Edberg joined the first field test for a new tool designed to break down scientific data silos: UAB Biomedical Research Information Technology Enhancement, or U-BRITE.
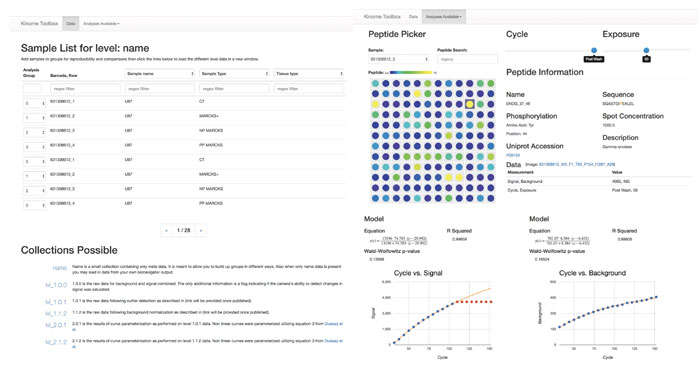 The interactive Kinome Toolbox was created by a UAB team led by radiation oncologist Christopher Willey. It lets users see the correlations among a host of data points, including gene sequencing, kinase activity profiles, tumor growth rates and treatment responses.
The interactive Kinome Toolbox was created by a UAB team led by radiation oncologist Christopher Willey. It lets users see the correlations among a host of data points, including gene sequencing, kinase activity profiles, tumor growth rates and treatment responses.
‘It even works on a cellphone’
U-BRITE gives investigators access to secure, high-volume storage for all the files that accompany a modern biomedical research project, along with a pipeline for clinical data and the high-performance computing resources to analyze all this information, explains James Cimino, M.D., director of the UAB Informatics Institute, which created U-BRITE, and co-director of the CCTS, which featured the project in its recent grant renewal. The U-BRITE framework is also designed to facilitate collaborative work among the investigators on a research team and interactions with the data scientists that support them.
In November 2018, after a six-month U-BRITE pilot, the lupus team demonstrated their progress along with three other groups of researchers and bioinformaticians from the UAB Informatics Institute. The lupus researchers had built the SLE Rosetta Database, a searchable platform connecting participant data. Rosetta allows them to visually analyze genetic variations seen by race and ethnicity, cluster recruited patients by genotype and visit time, and explore the availability of different clinical data in the study over time.
Radiation oncologist Christopher Willey, M.D., Ph.D., is exploring the signaling of protein kinases to develop new treatments for the brain cancer glioblastoma multiforme. He demonstrated an intuitive, web-based interface that lets users “see the correlations” among a host of data points, including gene sequencing, kinase activity profiles, tumor growth rates and treatment response. “And it even works on a cellphone,” said Willey, an associate professor in the Department of Radiation Oncology. Willey also recently received a UO1 grant with a computational aim that will further utilize the U-BRITE initiative.
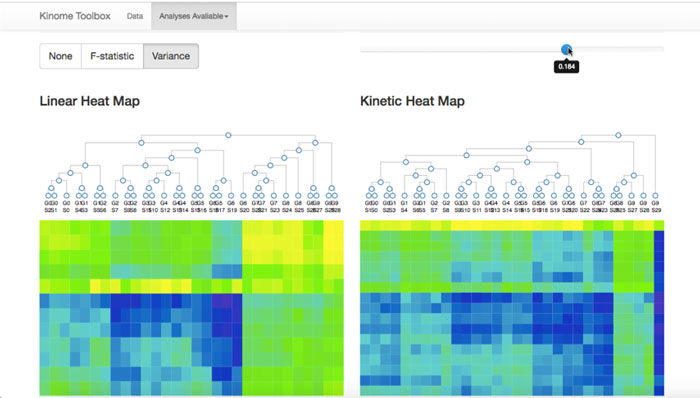 Another screenshot from the Kinome Toolbox illustrates the project's ability to visualize complex data.
Another screenshot from the Kinome Toolbox illustrates the project's ability to visualize complex data.
Everybody’s problem
“Everybody in the scientific community has similar problems and is clamoring for bioinformatics support,” Cimino said. The Informatics Institute provides that support to UAB researchers in collaboration with the CCTS. But each new project, with its unique aims and scientific questions, generally results in a custom solution. “If you want to ask a different question with the data, that doesn’t help you,” Cimino said.
The Informatics Institute and the CCTS envision a solution that could scale to empower team science across UAB and beyond. To test the U-BRITE concept, the Institute put out a request for proposals, seeking a variety of knotty, real-world research problems. In addition to the Edberg and Willey projects, the institute funded grants for teams with translational projects in rheumatoid arthritis (led by S. Louis Bridges Jr., M.D., Ph.D.) and precision genomics (led by Eddy Yang, M.D., Ph.D.) “We figured if we could solve their problems, we had solved problems for translational genomic/phenomic team science,” Cimino said. “And if we did it for several of them, it would be a generic solution, not a one-off.”
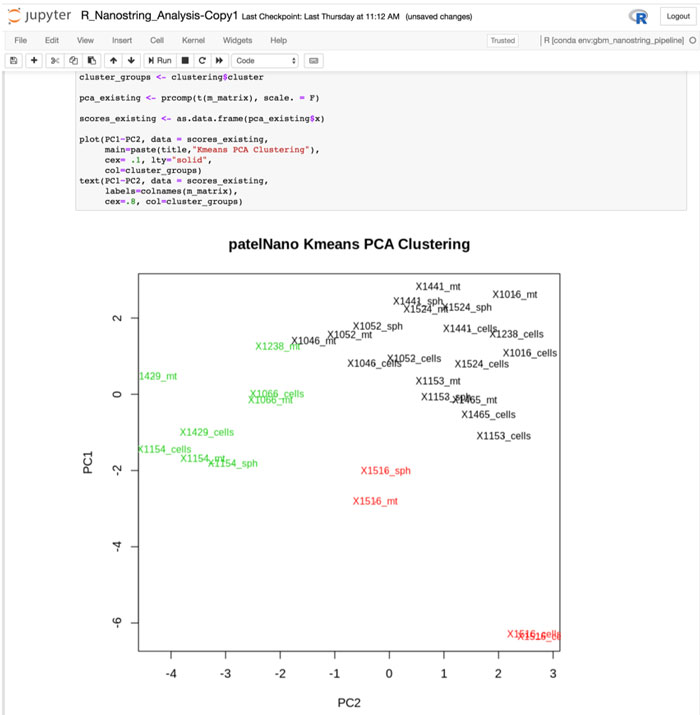 With Jupyter Notebooks, research teams can code collaboratively and see the direct connection between programming and visual representations of their data.
With Jupyter Notebooks, research teams can code collaboratively and see the direct connection between programming and visual representations of their data.
‘Easy and stress-free’
U-BRITE gives researchers a secure, HIPAA-compliant repository for data and includes a project management interface to wrangle masses of files, explains Jake Chen, Ph.D., UAB Informatics Institute chief bioinformatics officer and associate director and U-BRITE project lead. It also offers direct access to Cheaha, UAB’s supercomputer — one of the five fastest at academic institutions in the Southeast — for self-service analysis of next-generation genomic sequencing data. But beyond the technical offerings, U-BRITE also aims to demystify data analysis. “What we’re trying to do is provide an easy and stress-free data science environment to empower team science and drive advanced informatics method development,” Chen said.
To that end, U-BRITE includes access to Jupyter Notebooks, web-based applications running on the Cheaha supercomputer that let groups share live code and visualizations. Some coding is still required, “but that can be done by scientists in a principal investigator’s lab who have taken our courses,” Chen said. “The process is open so that investigators can understand how code produces graphs and other visualizations and they have more input. And if they get stuck, they can call in our bioinformaticians. We want to get people who are not traditional data scientists used to the idea that data science is not intimidating.”
Bringing everyone together
At the U-BRITE Day demonstration in November, Willey explained how Jupyter Notebooks give teams shared access to code and analyses that used to live “on one graduate student’s laptop,” because that was the only computer that could run it.
|
"The only way we were able to bring all this together was to bring everyone in the same room with U-BRITE." |
U-BRITE Day gave the research teams a chance to demonstrate what they had built in collaboration with the data scientists at the Informatics Institute. The Bridges team integrated data from i2b2, a self-service application that enables UAB researchers to access de-identified patient records, with the Rheumatology Arthritis Database and Repository (RADAR) maintained at UAB. That has allowed them to test correlations among genetic markers, disease severity and a host of patient traits, including gender, smoking status and the presence of CCP antibodies. The goal is to develop a model to predict radiographic joint damage in patients with rheumatoid arthritis in order to improve treatment.
Curtis Hendrickson, a UAB CCTS research associate and member of the Edberg team, demonstrated how they used Jupyter Notebooks and Cheaha to integrate genomic, kinomic and clinical data, along with biological data stored in UAB’s biobank. “The only way we were able to bring all this together was to bring everyone in the same room with U-BRITE,” Hendrickson said.
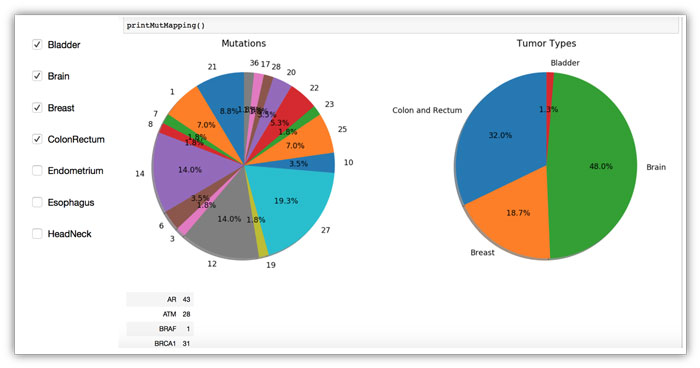 One of Eddy Yang's projects analyzed patient cancers sequenced at UAB by tumor type.
One of Eddy Yang's projects analyzed patient cancers sequenced at UAB by tumor type.
With U-BRITE, Yang’s team was able to store and analyze a vast collection of sequencing data from the 500-plus patient tumors sequenced at UAB through the STRATA trial. “Understanding the tumor genomic landscape at UAB can generate new avenues of future research,” said Yang, professor and ROAR Southeast Cancer Foundation Endowed Chair in the Department of Radiation Oncology. He demonstrated a dashboard interface allowing researchers to explore the distribution of mutations and tumor types for different subsets of cancer patients. He also demonstrated a proof-of-concept link between clinical data from i2b2 and the NIH’s ClinicalTrials.gov website, which in the future could “aid in identifying eligible patients for clinical trials,” he said.
Next steps
The Informatics Institute aims to incorporate lessons from the 2018 pilot into the next version of U-BRITE and add new features. A second pilot project, this time incorporating image data, may also be launched in conjunction with the CCTS—development plans for U-BRITE were favorably reviewed in the CCTS renewal grant.
The goal is to create “not just a piece of software, but a specification” that allows others to add other applications and adapt U-BRITE to new research problems and be disseminated to other universities, Cimino said. “Our mission is to expand the scope and scale of available data and knowledge resources to support team science and enhance the expertise of our research teams through training and collaboration. This isn’t a substitute for the science anyone is doing. We want to enhance it.”
For inquiries about how to participate in U-BRITE projects, contact Jake Chen at jakechen@uab.edu or visit ubrite.informatics.uab.edu/.