Benjamin Larimer’s project is focused on providing an extremely detailed look at the antibodies produced by people who have recovered from COVID-19. Could millions of viruses specializing in attacking bacteria offer new insights into SARS-CoV-2 — the virus threatening humanity in the coronavirus pandemic?
Benjamin Larimer’s project is focused on providing an extremely detailed look at the antibodies produced by people who have recovered from COVID-19. Could millions of viruses specializing in attacking bacteria offer new insights into SARS-CoV-2 — the virus threatening humanity in the coronavirus pandemic?
Benjamin Larimer, Ph.D., an assistant professor in the Department of Radiology and O’Neal Comprehensive Cancer Center at UAB, thinks so.
Larimer is one of 14 recipients of pilot funding through the School of Medicine’s Urgent COVID-19 Clinical Research and Laboratory Research Fund, which has raised more than $1.1 million in philanthropic support to fuel innovative research proposals. He is using that support to adapt his lab’s work on a technique known as phage display — which he normally uses to look for new cancer treatments — in order to make an impact on COVID-19.
Larimer’s project is focused on providing an extremely detailed look at the antibodies produced by people who have recovered from COVID-19. Antibodies recognize antigens — in this case, SARS-CoV-2 — but they don’t recognize the whole virus at once, Larimer explained. Instead, “they recognize a small piece,” he said. “The coronavirus is like a ball with a bunch of spikes all over it. The immune system generally recognizes things like that, so most antibody tests coming out now” are designed to look for antibodies in a person’s blood that bind to sections of that spike protein. “The problem is this is just one coronavirus; there are other types that have similar spikes — not extremely similar, but similar enough that you could potentially have antibodies against, say, a type of coronavirus that causes the common cold,” Larimer said. “And that would trigger a positive antibody test, even though you aren’t actually immune to the current SARS-CoV-2 coronavirus."
(Story continues below box.)
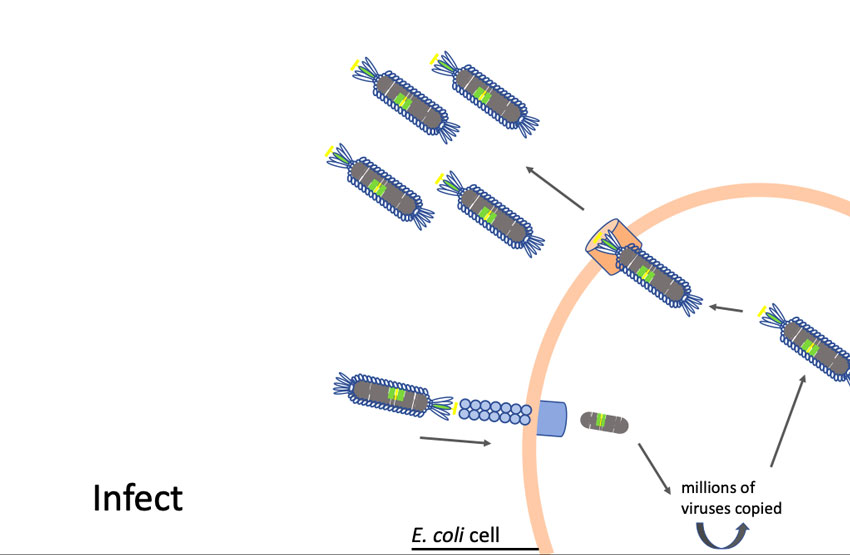
Phages are viruses that attack bacteria. Larimer trained at the University of Missouri with George Smith, Ph.D., who won a Nobel Prize in Chemistry in 2018 for his work in developing phage display as a way to evolve new proteins.
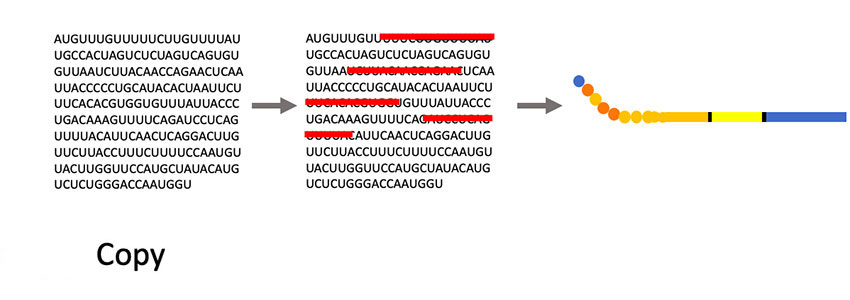
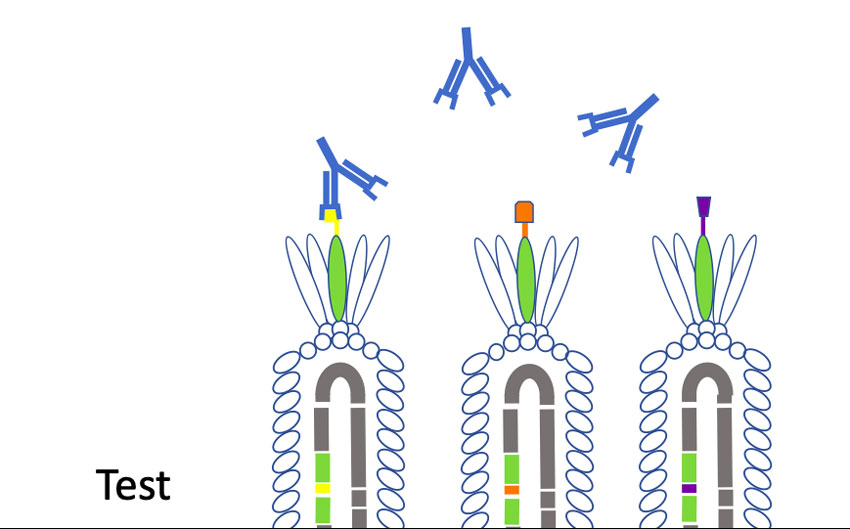
“Phages have this unique property where we can insert small pieces of DNA — for instance the DNA of coronavirus proteins, minus the proteins that allow them to be infective — and then read out which part of the protein is being displayed,” Larimer said. “My lab will make a huge library that contains all the non-infective proteins of the coronavirus. Then we’ll mix that library with a suspected antibody against the coronavirus that causes COVID-19. We’ll pull out the phage they are binding to; that phage contains the DNA sequence that tells us what exact piece of the coronavirus they are recognizing. It will allow us to more specifically define what you are immune to and overcome some of the limitations of current antibody tests.”
One real-world application, Larimer said, would be as a second-level test to verify results for those with positive readings on antibody blood tests. And if, as happened with coronavirus tests, PPE and ventilators, antibody tests become hard to source, “we wanted to be able to give UAB something that would help in day-to-day operations,” Larimer said.
| “Phages can be grown to high numbers in simple conditions. We can scale these up pretty inexpensively here at UAB and make the tests.” |
“The other interesting thing you could do with this is around vaccines,” he added. “The vaccines are attempting to induce immunity without exposing people to the virus. They are engineered in a lot of different ways, but many are using the spike protein. And the question that will emerge is, does the vaccine induce the same immunity as if you had acquired that immunity naturally by having COVID-19? We’ll be able to map, specifically, does this vaccine produce antibodies against the same pieces of the virus as we see in recovered patients, or against different pieces? If the first generation of vaccines is not as effective as we hope, we can say, ’Let’s use these common pieces of protein that we keep seeing in people who are naturally immune.’”
Phages also “are extremely inexpensive to produce,” Larimer said. “One of the limitations of current antibody tests is that you’re making a big spike protein, which takes protein-engineering expertise, materials, big reactors. Phages can be grown to high numbers in simple conditions. We can scale these up pretty inexpensively here at UAB and make the tests.”
That is not to say there is no challenge involved, Larimer added. “Probably the first time we test this in antibodies from patients we’ll have to tweak it,” he said. “It will take a few weeks to play with conditions and maximize specificity. My goal is to put this together, validate it on known antibodies, then test on blood samples from a subset of patients we know are COVID-positive and then on blood from before COVID-19 existed, so we know those are negative. Right now, we’re focused on is getting this ramped up and seeing how it looks.”
Related stories:

Coronavirus antibody testing now is available at UAB. Here’s what that means — and what it doesn’t.

Glow-in-the-dark coronavirus testing is first project of new hire

Researchers model COVID-19 infection in 3D human lung tissue

Tackling his third pandemic, UAB researcher gets up close with coronaviruses in order to kill them

Researchers are creating a coronavirus showdown to settle pressing antibody problems

Where are the good antibodies against COVID-19? UAB project aims to find out
Cytokine storm treatment for coronavirus patients is focus of first-in-US study

UAB launches research studies to combat COVID-19