No one wants to get coronavirus. Unfortunately, most everybody already has.
No one wants to get coronavirus. Unfortunately, most everybody already has.
Not the coronavirus, as in SARS-CoV-2, the novel coronavirus that causes COVID-19, but OC43 and HKU1, two other members of the same beta-coronavirus family. "Twenty percent of what we call colds are caused by these two coronaviruses that circulate widely in humans," said Troy Randall, Ph.D., professor in the School of Medicine departments of Medicine and Microbiology. "Most of us have been infected at least once or twice in our lifetimes by these two."
If tests to detect antibodies against SARS-CoV-2 in the blood pick up antibodies against the cold-causing coronaviruses instead, the results will be misleading at best. "When we find antibodies, how many of those were generated in response to a different virus and they are simply cross-reacting to places on SARS-CoV-2 with a similar shape?" asked Randall, who holds the Meyer Foundation, William J. Koopman Endowed Professorship in Rheumatology and Immunology at UAB.
"That's interesting in a scientific sense but really important in a public health sense," he said. "If many people have antibodies from a different virus that cross-react to SARS-CoV-2 it will look like they have immunity, but it's not actually immunity against the current virus. We need to understand how that works."
Building spikes
Randall and Frances Lund, Ph.D., professor and chair of microbiology at UAB, have spent decades studying antibody reactions to influenza. In a pilot project supported by the School of Medicine’s Urgent COVID-19 Clinical Research and Laboratory Research Fund, the researchers are adapting their proven methods to dig into the immunity question — one of the hottest topics in COVID-19 research.
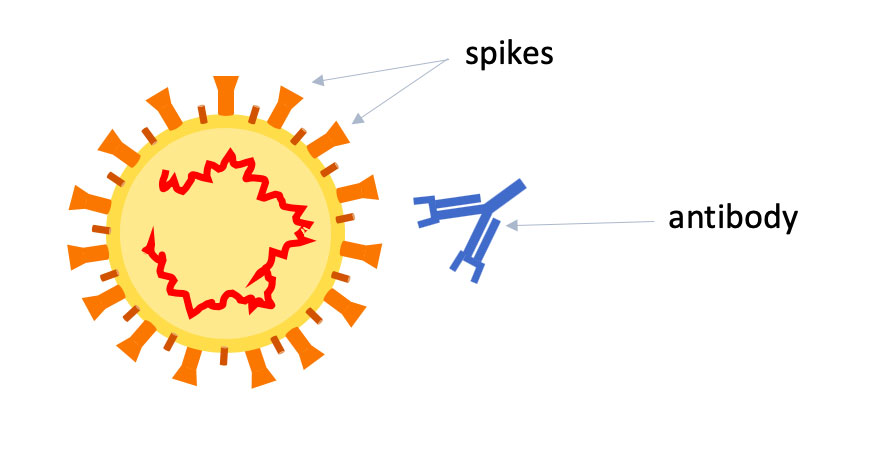
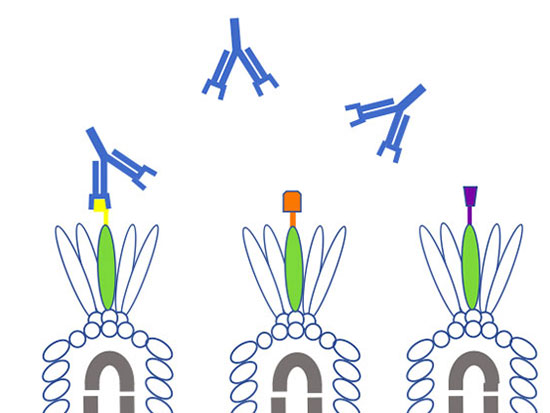
 Troy Randall, Ph.D.Lund and Randall will start by making two key proteins from five different beta-coronaviruses in their labs: the spike, or S, protein on the outside of the viruses, which is how they enter into cells, and the nuclear, or N, protein, which wraps up the RNA code inside the viruses. They will make S, N — and three different subsets of the S protein — for SARS-CoV-2, the original SARS, MERS and the "cold" viruses OC43 and HKU1.
Troy Randall, Ph.D.Lund and Randall will start by making two key proteins from five different beta-coronaviruses in their labs: the spike, or S, protein on the outside of the viruses, which is how they enter into cells, and the nuclear, or N, protein, which wraps up the RNA code inside the viruses. They will make S, N — and three different subsets of the S protein — for SARS-CoV-2, the original SARS, MERS and the "cold" viruses OC43 and HKU1.
"We've been working on immunity to influenza virus for decades, and we do exactly this same thing for flu," Randall said. "Every year the CDC and WHO come out with their best guess on what the circulating strains will be for the next year. Those strains go in that year’s vaccine. Once those are released, we make those proteins in the lab. Then, in people who are vaccinated or infected, we look in their blood at antibody titers and look at the B cells that make those antibodies."
How long is ‘immune’?
Why all the interest in pieces of S? The spike protein has been the focus of most vaccine and therapy efforts — particularly its receptor binding domain, which is the tip of the spike that interacts directly with human cells.
| "Twenty percent of what we call colds are caused by these two coronaviruses that circulate widely in humans. Most of us have been infected at least once or twice in our lifetimes by these two. When we find antibodies, how many of those were generated in response to a different virus and they are simply cross-reacting to places on SARS-CoV-2 with a similar shape?" |
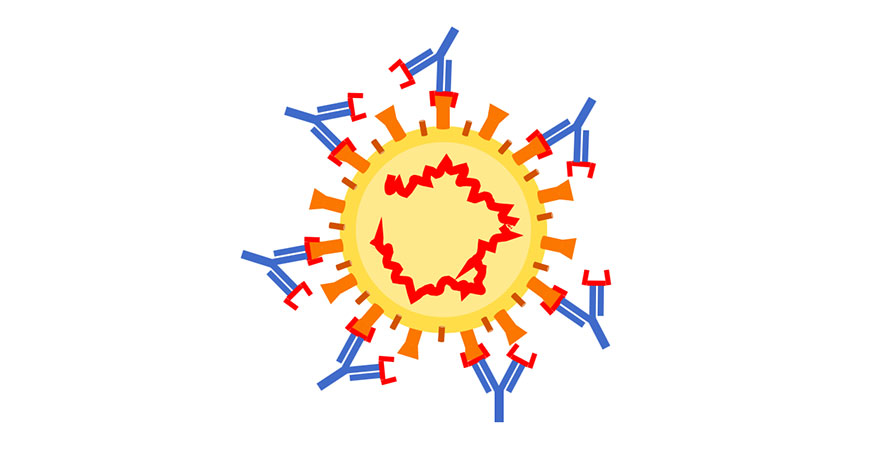
Even if the antibodies that appear on patient tests are, in fact, specific to SARS-CoV-2, it is important to understand which parts of the virus are being recognized. "Are they binding to the receptor-binding domain? That's probably really good," Randall said. "To the whole spike protein? That's probably pretty good. Right now we just don't know."
Another crucial question: How long do these SARS-CoV-2 antibodies last? Most people with functioning immune systems "develop antibodies against viral proteins as early as seven days from the onset of infection and these antibodies are detectable in serum for months, leading to a sustained and specific signature of prior infection," Randall and Lund noted in their application for the SOM's COVID-19 research program.
But "in the original SARS epidemic, immunity didn't last long — maybe a year," Randall said. "If you get it in 2020 and all your antibodies go away by 2021 that means you can be re-infected. Nobody wants that, but that's really important to know."
Rapid screen
 Fran Lund, Ph.D.
Fran Lund, Ph.D.
The goal of Randall and Lund’s project is to make “reagents that will allow us to rapidly screen for the presence of antibodies in the blood of acute and convalescent COVID-19-positive patients," said Lund, who is Charles H. McCauley Professor in the microbiology department. "We plan to use these reagents to assess how long the antibodies last in patients that were infected here in Alabama."
The difference between the tests that Lund and Randall are developing, and the antibody tests being used in hospital labs, including at UAB, is a matter of speed and precision. "A pathology lab can get hundreds of samples a day," Randall said. "They need a fast, reliable test that gives a definitive answer.” The test that he and Lund are developing is still pretty fast — it will take about an hour — but “is more complex because we're measuring different proteins from different viruses all at the same time,” Randall said. “Our test will tell you the amount of antibodies you have — whether they are N protein or S protein or certain parts of S. More antibodies is better, in general, and more antibodies against the receptor-binding domain is probably best."
But quality matters most. "You may have antibodies that stick to a protein, but do they actually neutralize the virus?" Randall said. "That is a more involved question."
| "If many people have antibodies from a different virus that cross-react to SARS-CoV-2 it will look like they have immunity, but it's not actually immunity against the current virus. We need to understand how that works." |
To find the answer, Randall and Lund are collaborating with Kevin Harrod, Ph.D., professor in the Department of Anesthesiology and Perioperative Medicine and UAB’s resident expert in SARS viruses. In Harrod’s lab, which operates at biosafety level 3, “with people in bunny suits and respirators and gloves,” Randall said, scientists deal with live samples of SARS-CoV-2. (In addition to Harrod, collaborators include Rodney King, Ph.D., John Kearney, Ph.D., and Todd Green, Ph.D., in Microbiology; James Kobie, Ph.D., and Paul Goepfert, M.D., in Infectious Diseases; and Marisa Marques, M.D., Jose Lima, M.D., and Sixto Leal, M.D., Ph.D., in Pathology.)
Neutralize them
"The assay is simple," Randall said. "You have a stock of the SARS-CoV-2 virus and you mix it with plasma from a patient who had the virus (or was vaccinated) and you add the mix to permissive cells. If the virus infects the cells, you don't have good neutralizing antibodies." The idea, Randall said, "is to figure out what aspects of our test most closely correlate with the neutralizing assay. Is it really the antibodies to the receptor-binding domain that are best correlated with protection?
"There is probably an antigen that will best trigger neutralizing antibodies," Randall said. "It's super important to figure out what that antigen is."
The researchers plan to compare their test with commercial tests to understand "if they give us the same information and are there drawbacks or advantages to one or the other," Randall said.
Lund and Randall are working with the Maryland-based pharmaceutical company Altimmune, which is developing a vaccine to prevent COVID-19. "We want to know — in vaccinated animals, or in people — are they making good antibodies, how long do they work and how long do they last?” Randall said. “Maybe you can make a better antibody response with vaccination. But we have to make these reagents first."
Read more on UAB's urgent COVID-19 research projects

Glow-in-the-dark coronavirus testing is first project of new hire

Researchers model COVID-19 infection in 3D human lung tissue

Tackling his third pandemic, UAB researcher gets up close with coronaviruses in order to kill them

Where are the good antibodies against COVID-19? UAB project aims to find out
Helpful viruses may unlock the secrets of coronavirus antibodies
Cytokine storm treatment for coronavirus patients is focus of first-in-US study