Shahid and Karolina Mukhtar in their Campbell Hall greenhouse with some of their thousands of Arabidopsis plants. You may not care who At4g31330 hangs out with, but odds are your grandchildren might.
Shahid and Karolina Mukhtar in their Campbell Hall greenhouse with some of their thousands of Arabidopsis plants. You may not care who At4g31330 hangs out with, but odds are your grandchildren might.
The world’s population is predicted to surpass 9 billion halfway through this century. To feed all those people, a group of British and American researchers wrote in a 2019 paper, “we need to increase world food production by at least 60% using the same amount of land, by 2050.”
The problem is worse than that, as the numbers show. By every indication, the climate and growing conditions will become increasingly harsh, shrinking crop yields through drought and increasingly acidic soils. Up to 30% of crops already fail each year, costing the global economy some $220 billion. Which leads us back to At4g31330, a so-far mysterious gene involved in protecting Arabidopsis thaliana, a member of the mustard family. Arabidopsis is worthless as a crop plant, unlike its relatives the cabbage and radish. But it plays a critical role in our future: Arabidopsis was the first plant to have its genome sequenced — even before the human one — in 2000. And because its genome is similar to that of many crop plants, Arabidopsis discoveries can be quickly translated into a boost in the food supply.
“We all eat plants,” said Shahid Mukhtar, Ph.D., associate professor in the Department of Biology. “We need to take swift action to increase crop production.”
With a new four-year, $1 million-plus grant from the National Science Foundation, Mukhtar and his research partner and wife, Karolina Mukhtar, Ph.D., associate professor and associate chair in the biology department, are doing something big. The researchers are using machine learning and other high-tech approaches to identify fresh ways to squeeze extra growing power out of the world’s crops. Then they will validate their discoveries in a lab packed with thousands of plants — and train dozens of Alabama high school science teachers to use simple experiments to help their students grasp the power of genetics.
Story continues below infographic

Decoding the ‘black box’ inside plants
Back in the early 2000s, when the Mukhtars were just beginning their doctoral training, they decided to focus their research on Arabidopsis. They were particularly interested in its response to heat, invading organisms and other stresses (Karolina) and in mapping the intricate pathways of gene and protein interaction that make up the plant’s immune system (Shahid).
“Yes, plants have immune systems. People sometimes have trouble believing that. But throughout their life cycle, plants are under attack and are constantly exposed to a wide range of pathogens.”
“Yes, plants have immune systems,” said Karolina Mukhtar. “People sometimes have trouble believing that. But throughout their life cycle, plants are under attack and are constantly exposed to a wide range of pathogens,” including viruses, bacteria, fungi, worms and oomycetes, also known as water molds. (The parasite responsible for the Irish potato famine was an oomycete.)
Like humans, Arabidopsis has about 30,000 genes. Those genes code for proteins, which interact with one another to form molecular machines, switch each other on or off, and more. In 2011, Shahid Mukhtar was lead author of one of two papers in the journal Science exploring the Arabidopsis “interactome” — a map of the molecular interactions inside Arabidopsis cells — like never before. In their paper, Shahid Mukhtar and colleagues analyzed interactions among more than 8,000 Arabidopsis proteins and two classes of effector proteins produced by common pathogens that attack Arabidopsis: a bacterium and an oomycete. The attacking effector proteins, Shahid Mukhtar reported, tended to target highly interconnected hub proteins at the center of interaction networks in Arabidopsis.
“Not every gene contributes equally to shaping plant behavior,” Shahid Mukhtar said. A biological network shares many features with social networks such as Facebook, he explains. “Every node in a network does not have an equal number of connections. Some people have 4,000 friends on Facebook, others may have only a few dozen. It’s the same in biological systems. Using such algorithms, now we can predict the host proteins that are targeted by pathogens.” (As Shahid Mukhtar and colleagues did in a 2018 paper in Nature Communications.) Protein interaction analysis gives important clues to which genes are worth studying further, he says.
The other Science paper back in 2011 described more than 6,200 interactions between some 2,700 proteins in Arabidopsis. That was impressive, but it represented only a subset of the genome. “Right now, 30% of genes in Arabidopsis are annotated as unknown,” Shahid Mukhtar said. For these 10,000-or-so genes, “it’s a black box,” he said. “They could be doing anything in the cell.” And even the 70% that are annotated may be incorrect or incomplete.
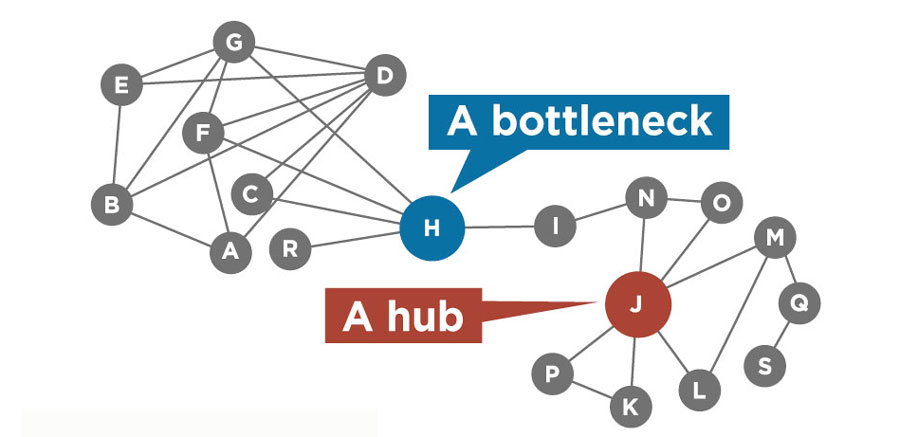 "Every node in a network does not have an equal number of connections," Shahid Mukhtar said. His research identifies the hubs and bottlenecks that are potential targets for pathogens and pests — and for defending plants against those pathogens.
"Every node in a network does not have an equal number of connections," Shahid Mukhtar said. His research identifies the hubs and bottlenecks that are potential targets for pathogens and pests — and for defending plants against those pathogens. Six degrees of Arabidopsis
The number of connections a protein has with other proteins is one key measure of its importance. “We have approximately 20 network features in three separate categories that we always analyze to figure out what components of the system are most important,” Shahid Mukhtar said.
“We know that acquisition of certain nutrients through the diet primes us humans to have a better immune response. We are trying to see if the same may be true in plants.”
With the new NSF grant, his lab is using two approaches: network topology and deep learning, a form of machine learning. “We are trying to feed a learning system millions and millions of combinations of transcriptome, interactome and other -omics data and see if it can learn patterns and predict function,” he said. “The concept has already been validated in our preliminary studies. We have a more than 90% success rate in correctly identifying gene function using our machine learning approach.”
Important genes and proteins are ones that should be the targets of future study; engineering these genes could make a plant more resistant to disease and other stressors. But how can researchers know for sure? That’s where Karolina Mukhtar’s lab comes in. “We both worked in wet labs in graduate school, but Shahid has moved to computational approaches, while I stayed loyal to pipettes and centrifuges,” Karolina Mukhtar said.

‘Amazon for plant biologists’
In particular, the Mukhtars are focusing first on genes implicated in processing of sulfur, a key plant micronutrient. “We know that acquisition of certain nutrients through the diet primes us humans to have a better immune response,” Karolina Mukhtar said. “We are trying to see if the same may be true in plants.”
“We are trying to feed a learning system millions and millions of combinations of transcriptome, interactome and other -omics data and see if it can learn patterns and predict function. In our preliminary studies.... we have a more than 90% success rate in correctly identifying gene function using our machine learning approach."
Sulfur is an important part of many biological models. “We have preliminary data showing that Arabidopsis mutants deficient in sulfur were not normal after infection,” Karolina Mukhtar said. Pathogens that attack plants are mainly after simple sugars, which they do not have to break down further. “But pathogens also try to siphon micronutrients such as sulfur and zinc as well, to fuel their growth and development,” she said.
The grant’s second aim is to “take at least 100 newly predicted sulfur genes, get knockout mutants [seeds with genes of interest inactivated, or “knocked out”] and subject them to a wide array of tasks,” Karolina Mukhtar said. These include growing the seeds in different stress conditions including drought and highly salty soil states predicted to result from climate change over the next few decades. This will be followed by biochemistry and molecular biology-related experiments. Genetic knockouts that result in hardier plants can then be investigated further.
“This is how in the long term we can identify plants that can survive in 2050, when the environment will be very different than it is today,” Karolina Mukhtar said. She has done this before. As a doctoral student, her research on the oomycete Phytophthora infestans, led to the development of a potato that is 11% more resistant to disease.
How do they get the knockouts? The Mukhtars could make them in their own lab. But a vast effort led by the Salk Institute in California has resulted in an online shopping site where researchers can order Arabidopsis seeds with virtually any possible gene inactivated, amplified or in other way mutated — for only a few dollars per seed. “It’s like Amazon for plant biologists,” Karolina Mukhtar said of the digital seed catalog, now maintained at Ohio State University. “You just search for what you need, click ‘add to cart,’ and in a few days the seeds are in our lab. And once you have a seed, you can grow thousands of plants from it.”
Growing a new generation of Alabama scientists
For the past several years, the Mukhtars have devoted part of each summer to teaching a genetics workshop for high school science teachers that they call Green DNA Day. One of the primary aims of their NSF grant is to transform those sessions into a three-week course series called Plant GIFT — Genomics Internship for Teachers.
“After the teachers train with us in the summer, they can take these lesson plans and experiments right back to their classrooms. The state can’t afford to have every high school class work on cancer cell lines in animals. But they can afford to give each class some seeds and let them see genetics in action.”
Starting in summer 2022 and focusing on teachers in northern Alabama and in rural counties, Plant GIFT will be “an intense immersion program, with lectures on theoretical material interspersed with hands-on lessons in growing plants and conducting experiments” on seeds that have one or more of the 100 identified sulfur genes knocked out, Karolina Mukhtar said. (Teachers will receive stipends of a few thousand dollars each for their participation.)
To simulate heat shock, for instance, the teachers may grow plants at 18-24 Celsius [64 to 75 degrees Fahrenheit], then raise the temperature to 42 degrees Celsius [107F] for 30 minutes, or 33 Celsius [91F] for an extended period. Over the next few days, the visible changes in the plants — wrinkled leaves and altered root structure, for example — will give them a dramatic picture of the effects of genetic variation. “Some mutants might be more tolerant, others will be less tolerant than the wild type,” Karolina Mukhtar said.
The Mukhtars are working closely with UAB’s Center for Community Outreach Development (CORD) and its director, J. Michael Wyss, Ph.D. (a professor in the Department of Cellular and Molecular Biology), as they develop the curriculum so that it aligns with Alabama’s new high school science requirements. “That way, after the teachers train with us in the summer, they can take these lesson plans and experiments right back to their classrooms,” Shahid Mukhtar said. “The state can’t afford to have every high school class work on cancer cell lines in animals. But they can afford to give each class some seeds and let them see genetics in action.”
Undergraduate and graduate biology students will play a major role in Plant GIFT, Karolina Mukhtar adds. Biology is the largest undergraduate major at UAB, and “we always have masses of students interested in doing research,” she said. “They are doing real work in our combined labs. And now our students will get an opportunity for valuable community engagement as they come with us to schools and showcase our work.”