-
What is COMET™?
What is COMET™?
The Lunaphore COMET™ platform is a fully automated liquid handler and microscope imaging scanner capable of performing sequential, multiplex, immunofluorescent staining and imaging samples adherent to a standard glass slide. The COMET™ system is fully customizable and capable of running 20 distinct staining conditions (i.e., optimization, mono-stains, dual-plexes) within a single run. The COMET™ system also operates with standard “off-the-shelf” antibodies, requiring NO conjugation. The transferability rate from applications optimized by hand or the BOND is very high. The COMET™ uses a proprietary microfluidic device that minimizes staining volumes and biochemistry dynamics, completing a single run in 24 hours. The platform has a four-slide capacity that maximally results in a 40-plex within 24 hours. Post-imaging, the slides can be used for multiple runs on the COMET™ or preserved for other applications (H&E, sequencing, inSitu Hybridization, thanks to the delicate elution buffer and low temperatures optimized for the sequential multiplexing).
For more information concerning the COMET™’s abilities and innovations please click here. -
Access To The Services
Access To The Services
The access to the services should be requested by sending a request to access to
fcsc@uabmc.edu
Please include the following information.
Full Name (First, Last):
Blazer ID:
Email:
PI full name.
The access is provided within 24 hours.
Once access to FBS has been provided please fill the COMET™ Sample Submission Form. -
Policies for COMET™ Services
Policies for COMET™ Services
- Assistance and Contact:
- All COMET™ runs are considered assisted services.
- Users are required to contact the Flow Cytometry and Single Cell Core Facility (FCSC) at harishpal@uabmc.edu to work out the COMET™ Usage Plan.
2. COMET™ Usage Plan
- Goal: to outline the optimal utilization of COMET™ staining platform and downstream analysis.
- Usage Plans will outline:
- Samples
- Submission form
- Sample preparation
- Quality Check
- Antibody Panels
- Addition of new antibodies
- Pricing
- Data handling
- Analysis plan
- Samples
- Samples
- Submission:
- All users must complete the sample submission form, providing comprehensive details about their samples.
- The form should include relevant information to facilitate the staining process effectively.
- Preparation:
- Investigators are responsible for preparing and positioning samples as per the guidelines provided by the facility.
- Proper preparation is crucial for optimal staining results.
- Quality Check:
- It is the responsibility of the users to verify the quality and positioning of their samples before processing.
- A preliminary analysis of H&E staining on serial sections is recommended.
- The area of interest should be within the imaging window on the COMET™, and indicated on the H&E stain/reference image.
- Users will be charged for slides if the staining protocol fails due to poor quality of tissue section or improper positioning.
- Antibodies and Panels:
- The antibody bank lists various antibodies and panels that have been tested and optimized on mouse or human tissues.
Requested antibodies/panels will be tested and optimized on investigators' samples before processing the full cohort for staining.
- Addition of New Antibodies
- The addition of investigator’s choice of a new antibody to existing panels requires optimization for staining conditions.
- Staining Assessment and Optimization:
- Upon completion of staining on the investigator's optimization slide, data will be transferred to the virtual computer drive or FCSC LTS to be retrieved for labs not using the Virtual Image Analysis Resources.
- Investigators are required to assess staining specificity and patterns promptly.
- Recommendations for changes in the staining protocol should be communicated as soon as possible for further optimization or running on the rest of the slides.
- Once the staining protocol is finalized and applied to all requested slides, data will be transferred to the virtual computer drive for user access.
- Data Handling
- User Responsibility for Data Management:
- UAB-FCSC is not responsible for the storage or management of user data for an extended period.
- Users must adhere to NIH data management guidelines.
- It is the user's responsibility to back up and store data appropriately.
- Users must inspect every image file for corruption within 14 days of delivery. Any requests for data re-delivery after 14 days may not be able to be met.
- Data Size and Periodic Removal:
- Multiplexed staining of a single slide generates a large data file (approx. 100 GB of data/slide for a 20 antibody panel) depending on the staining protocol.
- Once confirmed by the user for full accessibility (within 14 days of delivery), data will be periodically removed from the COMET™ computer to free up space for further use.
- Analysis
- Image Analysis resources are provided including:
- Computing Hardware through Virtual Access on Azure Computers
- Horizon Viewer
- Visiopharm Analysis Software
- Software Training
- Support for analytical design and execution
- Azure Core Computing Systems
- The Azure computers (2) are highly equipped data analysis computers with substantial CPU and GPU hardware, 30TB of shared storage and pre-installed analysis software.
- Limited Computer Storage:
- Users share limited computer storage space.
- Data can only reside on the virtual computer drive during analysis and must be removed upon project completion.
- Extended storage on the virtual computer drive will result in additional charges determined by the FCSC.
- Computing Time Reservation:
- Users are required to reserve computing time on the Facility Booking System (FBS) to access the UAB Azure Image Analysis Computer – The virtual computer.
- Reservation details and guidelines can be found on
- Logout Procedure:
- Users should be aware that closing/disconnecting from the Citrix server alone or allowing it to time out does not constitute a proper logout.
- Proper logout involves closing all applications and logging out of the windows user from the start menu
- Users are responsible for ensuring all analyses and software are closed before logging out to free up resources for others.
- Users who do not adhere to the logout procedures will be charged accordingly.
- Visiopharm Analysis Software
- Visiopharm is high end image analysis software specialized for histopathological image analysis including COMET™ mIF images.
- Interface Usage:
- Upon completion of the reservation time, users must Sign Out and actively close the Visiopharm user interface.
- Failure to properly sign out and close Visiopharm blocks the seat for other users and may result in charges
- Access Request and Training:
- Users must request access to the necessary image analysis software through email to harishpal@uabmc.edu or jcarstens@uabmc.edu
- It is the user's responsibility to ensure they have access to the required software and undergo necessary training for staining assessment and analysis.
- Training Resources:
- Image analysis training resources, including videos, are provided by Visiopharm on the Customer Portal (https://support.visiopharm.com/signin). Access must be requested by email to harishpal@uabmc.edu or jcarstens@uabmc.edu
- Recordings of onboarding training sessions, instructions for use of UAB provided “canned apps” are also available on the FCSC – Visiopharm Kaltura channel (https://mediaspace.uab.edu/channel/Visiopharm%2BTraining/310244572).
- Two one-hour bi-weekly shared Visiopharm office hours are provided. Recordings are made available on the FCSC UAB Box.
- Globus Transfer:
- COMET™ images are large and prone to corruption using standard windows explorer tools.
- Globus is a web-based transfer system provided by UAB to allow easy and secure data transfer with bit checks and is the recommended method for data transfer, backing up, etc.
- Provided nodes: UAB Azure node for Visiopharm Core computers, UAB Box, UAB LTS, and personal computing systems through Globus connect. Image analysis training resources, including videos, are provided by Visiopharm on the Customer Portal (https://support.visiopharm.com).
- Image Analysis resources are provided including:
- Charges for use of COMET services and Virtual Image Analysis Resources
- Users will be billed according to the number of antibodies used in the staining panel and number of slides used for staining as described in the “Charges for COMET™ Services and use of Virtual Image Analysis Resources” page and above in the polices.
- Users will be billed for use of Virtual Image Analysis Resources based on their monthly or yearly choice and is linked to data occupancy and/or FBS reservations.
- Users are responsible for properly signing out from Visiopharm, Azure Virtual Computer and any other software used on the Virtual Image Analysis Resources. Failing to properly sign out from these resources will result in full charge for whole period until signed out properly.
-
Image Analysis Resources
Image Analysis Resources
If your laboratory does not have your own independent access, all COMET™ users will need the FCSC Virtual Image Analysis Resources. To utilize the FCSC Virtual Image Analysis Resources, all users will be charged $370/month or $3,700/year. Monthly charges will be linked to data occupying hard drives and/or FBS scheduler bookings.
FCSC Virtual Image Analysis Resources include: Azure computing, active use Azure hard drive space, Horizon Viewer, FIJI, BFTools, and 30 hours/month of Visiopharm image analysis software access and support (bi-weekly office hours, training videos and canned apps for general UAB COMET™ data).
Neighborhood Analyses: The spatial analysis of COMET™ data will be available after the COMET™ images are analyzed with Visiopharm and the cell segmentation and phenotyping data are properly exported according to the standardized formats. Neighborhood analysis based on L- and G-functions that examine neighborhood relationships between cell or biomarker types have been developed within Immunology Institute and will be made available to the PI along with instructions and manuals. The PI may use the provided analytic tools to perform spatial analysis to acquire a standard set of features that measure the spatial correlation and co-occurrence patterns. For more advanced analysis, including more complicated biological interactions or advanced algorithms, customized development of analytic tools is available under the collaboration with faculty members in Department of Radiology (contact Dr. Dean Fang, yfang@uabmc.edu) and additional fees may occur for such demands.
Advanced Analytics Support:
- 4 hrs of one-on-one support from Visiopharm expert tech support - $1,540
- 8 hrs of one-on-one support from Visiopharm expert tech support - $2,420
- Advanced analytic support with customized coding and development - $150/hour
-
Services - Validated Antibody Bank and Optimized Panels
Services - Validated Antibody Bank and Optimized Panels
- All runs of the COMET™ system are required to use the FSCS COMET™ Antibody Bank. This invaluable resource accelerates our progress by allowing our community of users to build upon each other’s success and expertise.
- Currently, the bank represents panels reactive to human, mouse, rat, and canine tissue sections with markers spanning the tumor microenvironment, neurology, and immunology.
- This bank is a continually expanding resource and is guided by the experimental needs of our users.
- See the most up-to-date antibodies here and panels here. Please contact
fcsc@uab.edu should you have questions.
-
Services - Assisted Optimizations
Services - Assisted Optimizations
- All COMET™ runs are Assisted.
- Optimization of markers takes on two forms: Mono/single antibody and Multiplex.
- Mono-optimizations.
- This is for new antibodies being added to the bank or testing of antibodies already existent in the bank but not yet tested on the desired tissue type or organism.
- The detailed optimizations are broken down into 3 “characterizations” as guided by Lunaphore. The FCSC core has procured normal C57Bl/6 and human tissue arrays for this initial characterization. The requesting investigator must provide tissue types not reflected in this array.
- Characterization 1: Does it work?
Best guess concentration (usually on the high side) on tissues where it should express. - Characterization 2: What’s the best concentration/exposure?
Approximately 3 concentrations to pick the best exposures and concentrations. - Characterization 3: How stable is the epitope?
Initial stain, then serial elutions (5-20x) followed by identical staining.
- Multiplex optimizations.
This is for the optimization of a full multiplex panel of antibodies in the bank and has proven to work on the tissue of interest on your specific tissue of interest. Every tissue stains differently; some markers may need to be tweaked on new samples. This is a check before a full cohort is run. When successful, the exact staining conditions will be used on the multiplex panel experiment. All data is saved and can be used as one of the samples.
-
Rates and Fees
Rates and Fees
Charges for COMET™ Services and use of Virtual Image Analysis Resources
General Guidelines:
Users will be charged on a per slide basis depending on the number of antibody conditions that are used (See current Antibody Bank). Batch processing of fully optimized panels are run at a discounted instrument fee (1 fee across up to 4 slides run). See details below
Optimizations:
All new antibodies, new panel combinations, or new tissues using existing antibodies (including TMA if original optimizations were run on full sections) must undergo optimization. Users will be charged on a per slide basis depending on the number of antibodies that are used. Each optimization run is charged for instrument and technician time without the batch slide discount that is provided in optimized/Full runs table below.
All antibodies are purchased by the core. Failed clones (not added to the bank) will be charged in full to the investigator and can be collected for other laboratory uses.
New antibody validation:
New antibodies must be validated (confirmed by the investigator to express where expected) and titrated prior to inclusion in an experiment. The Core will purchase previously untested antibodies according to user recommendations and will validate/titrate those antibodies on positive and negative control slides provided by the user.
For example, a new antibody could be used at 3 concentrations on a positive control slide along with a panel of previously validated antibodies. Users would be charged for the run and for the number of antibodies (new antibody used at 3 dilutions counts as 3 antibodies) according to the pricing schedule below.
In another example, a user might want to validate multiple antibodies on a positive control slide. Each antibody would be tested at 3 concentrations. Thus, if 5 new antibodies were tested at 3 concentrations each, the charge for the test panel would be based on 15 antibodies and a charge for the run according to the schedule above.
Consultations with Harish Pal (harishpal@uabmc.edu) are required prior to submitting validation request forms to develop an optimization plan and a quote.
Optimized/ Full Runs:
Users will be charged on a per slide basis depending on the number of previously optimized antibodies that are used. (See current Antibody Bank)
The core applies a batch run discount to the instrument fee of up to 4 slides of the same panel in a single run. Instrument fee is $470/run. A run is completed in about 24-36 hours and can be run on a maximum of 4 slides.
For example, if a user wants to image 7 slides using 8 antibodies, the price will be (7 x $560/slide) + (2 x $470/run). Data collection will be completed after 2-3 days depending on instrument and technician availability.
Please consult with Harish Pal (harishpal@uabmc.edu) prior to scheduling the experiment so that a quote can be developed.
All charges listed in the table below are per slide used.
No. of antibodies/slides
1 slide
2 slides
3 slides
4 slides
1-5 antibodies = $430/slide + $470/run
$900
$665
$587
$548
6-10 antibodies = $560/slide + $470/run
$1,030
$795
$717
$677
11-15 antibodies = $690/slide + $470/run
$1,160
$925
$847
$807
16-20 antibodies = $820/slide + $470/run
$1,290
$1,055
$977
$937
>20 antibodies within a single run = $26/antibody/slide/concentration + $300 in consumables/slide + $470/run
For example, 30 antibodies/slide should be (30 x $26 = $780/slide for antibodies) + ($300/slide for consumables) = $780 + $300 = $1080/slide. Add $470 instrument fee/4 slides ($1,550/slide if you ran 4 slides).
Novel panels that require more than one run will be charged accordingly.
Data Handling and Virtual Image Analysis Resources:
To utilize FCSC Virtual Image Analysis Resources, all COMET™ users will be charged $370/month or $3,700/year. Monthly charges will be linked to data occupying hard drives and/or FBS scheduler bookings.
FCSC Virtual Image Analysis Resources include: Azure computing, active use Azure hard drive space, Horizon Viewer, FIJI, BFTools, and 30 hours/month of Visiopharm image analysis software access and support (bi-weekly office hours, training videos and canned apps for general UAB COMET™ data).
Neighborhood Analyses: The spatial analysis of COMET™ data will be available after the COMET™ images are analyzed with Visiopharm and the cell segmentation and phenotyping data are properly exported according to the standardized formats. Neighborhood analysis based on L- and G-functions that examine neighborhood relationships between cell or biomarker types have been developed within Immunology Institute and will be made available to the PI along with instructions and manuals. The PI may use the provided analytic tools to perform spatial analysis to acquire a standard set of features that measure the spatial correlation and co-occurrence patterns. For more advanced analysis, including more complicated biological interactions or advanced algorithms, customized development of analytic tools is available under the collaboration with faculty members in Department of Radiology (contact Dr. Dean Fang, yfang@uabmc.edu) and additional fees may occur for such demands.
Advanced Analytics Support:
- 4 hrs of one-on-one support from Visiopharm expert tech support - $1,540
- 8 hrs of one-on-one support from Visiopharm expert tech support - $2,420
- Advanced analytic support with customized coding and development - $150/hour
-
Sample Preparation
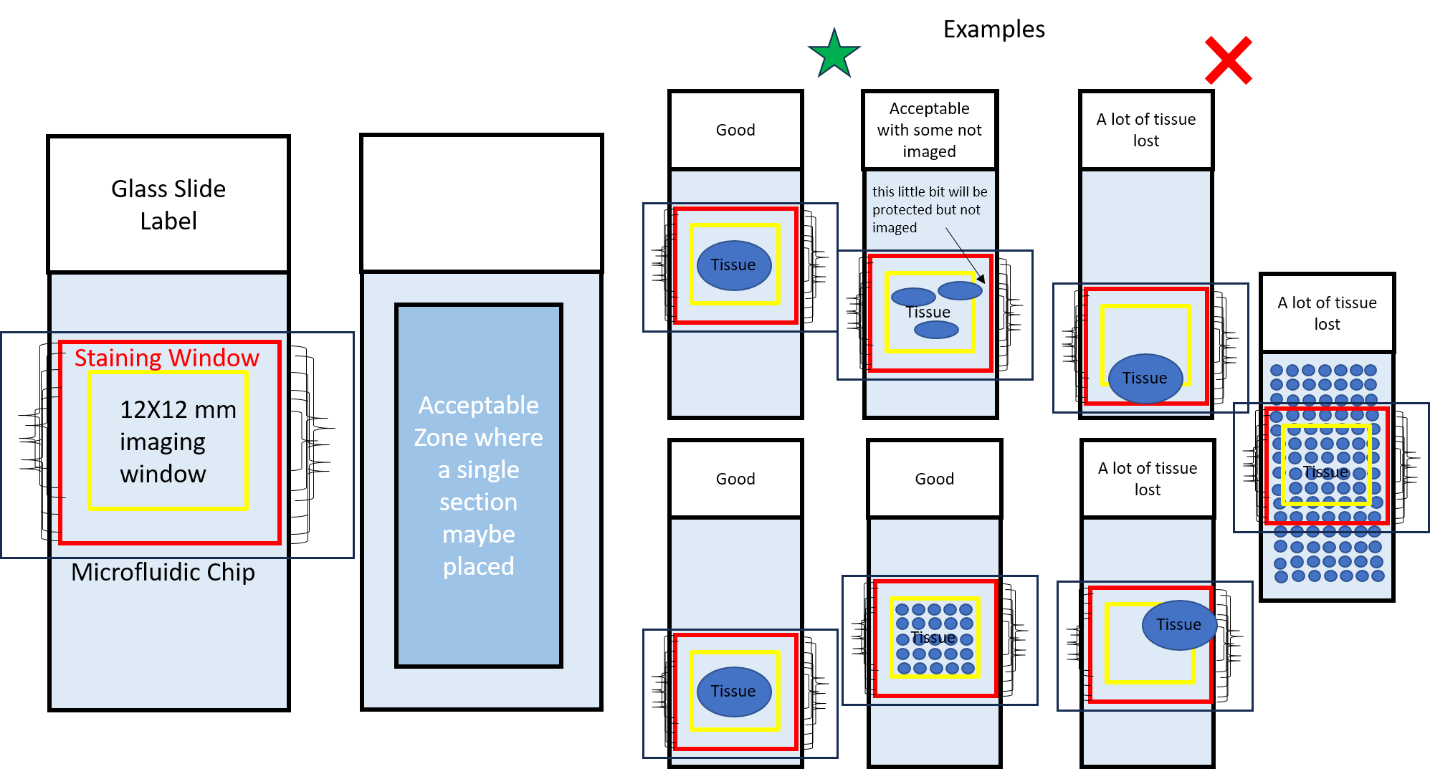
Sample Preparation
Sample can be anything that can attach to a standard microscopic slide and fit within the staining and imaging window of the microfluidic chip. Some examples are tissue sections, FFPE, snap frozen tissue, etc.
Where you place your sample matters. A model drawing of a slide is provided in figure 1. Anything within the blue box is an acceptable location for your sample. Thus, stars can be seen within the blue box as stars represent acceptable locations for your sample. In contrast, the triangles are located outside the blue box because they represent unactable locations for your sample.
The red square, in figure 1, represents a metal wire contained within the chip. Anything within this metal wire is safe.
The imaging window has the size of 12x12 mm. This imaging window will be directed toward the center of the slide. Thus, ensure that whatever you want to imaged is located at the center of your slide and can be contained within the 12x12 mm imaging window. Whatever part of your sample that is outside the 12x12 mm imaging window WILL NOT BE IMAGED. In figure 1, the 12x12 imaging window is represented by the yellow square.
If your laboratory does not have tissue sectioning capabilities, we have identified two facilities on the UAB campus that offer these services: Pathology Core Research Lab and the Animal Resource Program Comparative Pathology Lab.
-
NIH Grant Document
NIH Grant Document
Multiplex Immunofluorescence (mIF)
Three research institutes at UAB (O’Neal Comprehensive and Cancer Center, Immunology Institute, and I-4ward fund) have demonstrated continuing support in the form of physical infrastructure, personnel expertise (dedicated staff in a shared resource facility), and financial investment into the support of the proposed multiplex immunofluorescent (mIF) imaging studies. This combined investment has resulted in the recent purchase of the COMET™ multiplex imaging system, the establishment of an optimized antibody bank, and computational support for the analysis of mIF images in collaboration with the Research Computing and Cheaha Cluster teams. These endeavors are run at or below cost (i.e., supported by other grant mechanisms) to maximize the impact, accessibility, and successful use of these technological investments. Detailed training and workflows have been generated and are made available on-demand to all UAB and collaborating researchers. These investments and the collaborative interactions between the Biospecimen Bank (clinical samples) and Animal Core (pre-clinical samples) demonstrate an ideal environment for completing the proposed aims.EQUIPMENT
Lunaphore COMET™ Multiplexing System
Located within the Flow Cytometry and Single Cell Shared Resource on the UAB campus in the Shelby Research building. The COMET™ system is a self-contained liquid handler and microscopic image scanner in one fully modifiable program to optimize and validate three fluorophores each cycle. An optimized antibody bank and a BioGenex EZ-Retriever antigen retriever system support the COMET™ system. The fully trained core staff supervises all reagents, antibodies, protocol optimizations, and equipment operations.Visiopharm Image Analysis Software
UAB investigators and core facilities have access to several seats of the full research license of Visiopharm analysis software. Access to 75TB of storage for each research faculty within the Research Computing core is provided to store multiplex scanned images for analysis. These systems come with several hours of detailed user support and training each month and access to on-demand, online overview training for all future and current users, supported by Dr. Carstens.