During the past several decades, a string of highly promising therapies to treat Alzheimer’s disease have failed in clinical trials. Thousands of patients and families, at UAB and other trial sites around the world, have seen their hopes dashed. Researchers, who saw years of work and patiently elaborated hypotheses crumble, were devastated, too. But Richard Kennedy, M.D., Ph.D., suspects the wreckage of these failed tests may yet hold hidden clues.
 Richard Kennedy, M.D., Ph.D., is using machine-learning techniques to search for new Alzheimer's disease therapies among the 10,000 medications being taken by patients who participated in failed trials. In 2017, with a multi-year, multimillion-dollar commitment from the National Institute on Aging and a database of some 8,000 Alzheimer’s drug trial participants going back 30 years, Kennedy began testing a novel idea. He knows exactly how quickly the disease progressed in each of those participants. And he knows which medications (other than the test therapy) they were taking — drugs for heart disease, high blood pressure, diabetes and on and on. “That’s 10,000 or so medications that could be associated with slower progression of Alzheimer’s disease,” said Kennedy, an associate professor in the Division of Gerontology, Geriatrics and Palliative Care. “It’s a huge natural experiment.” The study participants’ primary care doctors “put them on these medications — not for Alzheimer’s, but they were taking them when they came into the Alzheimer’s study,” Kennedy said. And because “you get these cognitive assessments all along the way,” it’s possible to see if any of those medications was associated with better assessment scores.
Richard Kennedy, M.D., Ph.D., is using machine-learning techniques to search for new Alzheimer's disease therapies among the 10,000 medications being taken by patients who participated in failed trials. In 2017, with a multi-year, multimillion-dollar commitment from the National Institute on Aging and a database of some 8,000 Alzheimer’s drug trial participants going back 30 years, Kennedy began testing a novel idea. He knows exactly how quickly the disease progressed in each of those participants. And he knows which medications (other than the test therapy) they were taking — drugs for heart disease, high blood pressure, diabetes and on and on. “That’s 10,000 or so medications that could be associated with slower progression of Alzheimer’s disease,” said Kennedy, an associate professor in the Division of Gerontology, Geriatrics and Palliative Care. “It’s a huge natural experiment.” The study participants’ primary care doctors “put them on these medications — not for Alzheimer’s, but they were taking them when they came into the Alzheimer’s study,” Kennedy said. And because “you get these cognitive assessments all along the way,” it’s possible to see if any of those medications was associated with better assessment scores.
This is a pattern-matching problem on a vast scale — akin to finding matching stalks of hay in a colossal haystack. To do that, you would need to compare each strand to every other strand, across multiple dimensions. This is an ideal use case for machine learning — the subfield of artificial intelligence that has already revolutionized image search, fraud detection and many other applications. Machine learning is catching on with scientists, too. Between 2015 and 2018, the number of biomedical papers involving machine learning jumped to more than 8,000 per year from less than 1,000. Machine learning is at the core of Kennedy’s approach to Alzheimer’s and a second NIH R01 grant he received in 2019. In fact, Kennedy was one of three UAB researchers to receive significant NIH funding for machine-learning applications in clinical care this past year. Here, they discuss their work and the ways in which machine learning could revolutionize treatment for millions.
Finding the hay in a haystack
Just down the street from Kennedy’s lab, Brittany Lasseigne, Ph.D., is chasing another haystack issue. Lasseigne, a recipient of the NIH’s prestigious K99/R00 Pathway to Independence award, was recently recruited to UAB from HudsonAlpha Institute for Biotechnology in Huntsville (see our recent stories on other K99 recipients here, here and here). She runs a hybrid lab; there is a gene-sequencer in one corner, a high-powered computer in another and plenty of beakers, tubes and other typical wet-lab glassware in between. Her goal: to perfect “cell-free” sequencing tests that can track DNA and RNA shed from tumors and other dysfunctional cells — all in a single milliliter of patient blood. “Our question is, How can we learn something new about how a disease starts or how it progresses?” said Lasseigne, an assistant professor in the Department of Cell, Developmental and Integrative Biology. “Our inputs are genomic datasets — DNA, RNA or how they interact with proteins — and our outputs are biological signals or clinical outcomes. And we use machine learning to look across those datasets to find patterns. That lets us identify sets of biomarkers, so when we see those patterns in the future we know how a patient will respond.”
| “When you have a set of records where you know the classification, such as whether someone became delirious or not, especially if it’s a very large dataset where there are many possible predictors, that’s an indication that machine learning may be a good choice.” |
Meanwhile, one block away, Surya Bhatt, M.D., Ph.D., is looking for patient patterns of a different sort. A pulmonary physician specializing in chronic obstructive pulmonary disease (COPD), he is training a machine-learning model to pore over CT scans and recognize an often overlooked and untreated subset of patients. His work, which is funded by an R21 Exploratory/Developmental Research grant from the NIH’s National Institute of Biomedical Imaging and Bioengineering, “could lead to a paradigm shift in the field,” potentially improving treatment for millions, said Bhatt, an associate professor in the Division of Pulmonary, Allergy and Critical Care Medicine and director of the UAB Lung Imaging Core.
What is machine learning, anyway?
“There are a lot of debates about what’s machine learning and what’s not,” Lasseigne said. “If the computer is helping me with the equation, I call it machine learning.” It’s just that the help is a little different from what we’re accustomed to when using machines. “I think of it this way: A calculator takes a set of numbers — say 2 and 3 — and an operator — the ‘plus’ sign — and gives you 5,” Lasseigne said. “Machine learning turns that on its head. You give it the inputs 2 and 3 and the output 5 and ask the computer to help calculate it.”
| “There are a lot of debates about what’s machine learning and what’s not. If the computer is helping me with the equation, I call it machine learning.” |
That, of course, is an equation you can solve in your head. But try this instead: Choose 20-30 words at random out of the 10,000 or so in an average electronic medical record, then look through more than 50,000 medical records to see if they have any correlation with that patient’s eventual delirium status during a hospital stay. Then do it again, and again, until you’ve covered the whole lot. That’s the topic of Kennedy’s latest grant, another multi-year, multi-million dollar R01 from the National Institute on Aging using machine learning.
Investing in computational power
“When you have a set of records where you know the classification, such as whether someone became delirious or not, especially if it’s a very large dataset where there are many possible predictors, that’s an indication that machine learning may be a good choice,” Kennedy said. “This type of discovery process is very difficult using traditional tools. With a desktop computer it would take years.” But a multi-million dollar institutional investment in UAB Research Computing resources during the past several years means Kennedy now has the power to run this type of experiment on UAB’s 4,000-plus-core Cheaha supercomputer in a few hours or days. “Cheaha is a lot of what makes this possible,” Kennedy said. “We’re fortunate at UAB that we have a really good resource in our Research Computing department.”
Kennedy has worked closely with Research Computing staff such as William Monroe, who says he has been partnering with a growing number of UAB researchers during the past year from the schools of Medicine, Engineering, Dentistry and more.
Kennedy also has tapped into the expertise of the UAB Informatics Institute, including John Osborne, Ph.D., who specializes in natural language processing (see Watching his words, below). “You have to be able to pick up on the different way things are phrased in a patient chart,” Kennedy said. A doctor might write “patient has altered mental status,” while a nurse’s comment might say a patient was “acting weird.” “That’s not a diagnosis of delirium, but it can be an indicator of it,” Kennedy said. And, by the way, the computer needs to understand that “patient has altered mental status” and “patient’s mental status is altered” refer to the same thing, while “patient’s mental status is not altered” is the opposite.
What you can do at UAB
The university’s collaborative atmosphere was a key element in Lasseigne’s recruitment. “UAB is very team-science-oriented,” she said. “I can go have coffee with a clinician in the Cancer Center and talk about problems they’re seeing in patients — that is something you can only do at a major medical center.”
UAB also is willing to invest in innovative thinking, Bhatt said. His project grew out of his time in the Deep South Mentored Career Development scholars program in the UAB Center for Clinical and Translational Science. The program, also known by its NIH grant designation, KL2, is designed for junior faculty with a passion for translational research. Bhatt went on a CCTS-funded sabbatical to Auburn University to acquire new skills in cardiac MRI imaging with a biomedical engineering faculty member at Auburn. In the process, he met a visiting professor from Georgia Tech University who was an expert on fluid mechanics in the heart. “That was the genesis for this idea to study fluid mechanics related to major airway collapse in COPD,” Bhatt said.
Learn more about how Bhatt, Lasseigne and Kennedy are tackling their problems with computing power:
1. Finding confused patients before it’s too late
 More than 50,000 patients age 65 and older have received delirium screenings at UAB Hospital as part of the innovative Virtual Acute Care for Elders Unit program. With a machine-learning algorithm, Richard Kennedy (not shown above) is mining these records to find patterns that could allow doctors to predict which patients are at highest risk for delirium.The project: Automating Delirium Identification and Risk Prediction in Electronic Health Records, launched in spring 2019 with a $403,449 grant from the National Institute on Aging. Principal investigator: Richard Kennedy, M.D., Ph.D. Investigators: Kellie Flood, M.D., John Osborne, Ph.D.
More than 50,000 patients age 65 and older have received delirium screenings at UAB Hospital as part of the innovative Virtual Acute Care for Elders Unit program. With a machine-learning algorithm, Richard Kennedy (not shown above) is mining these records to find patterns that could allow doctors to predict which patients are at highest risk for delirium.The project: Automating Delirium Identification and Risk Prediction in Electronic Health Records, launched in spring 2019 with a $403,449 grant from the National Institute on Aging. Principal investigator: Richard Kennedy, M.D., Ph.D. Investigators: Kellie Flood, M.D., John Osborne, Ph.D.
The problem: Delirium, marked by confused thinking and a lack of environmental awareness, is extremely common in hospitals. According to a 2012 study, up to 25% of patients ages 65 and older who are hospitalized have delirium on admission, and another 30% will develop delirium during their hospital stays. But the true numbers may be far higher. “Delirium is not well recognized,” Kennedy said. “Research studies in hospitals found that 75% of the time, doctors were unaware their patients had delirium.” Identifying delirium is very important, he added. “It is associated with a higher risk of death and dementia.”
The data: For the past several years, every patient 65 and older admitted to UAB Hospital has received a delirium screening on admission and every 12 hours during their stay. It’s an idea borrowed from the hospital’s innovative Acute Care for Elders (ACE) Unit, which pioneers new approaches to improving care and quality of life for older patients. More than 50,000 patients have received screenings as a part of this Virtual ACE Unit program.
Kennedy is leveraging those 50,000 patient records, which give him an accurate picture of which patients did and didn’t receive a delirium diagnosis during their stay. Other researchers have attempted to isolate delirium cases by searching medical records for some two dozen keywords identified by experts. “They did terribly,” Kennedy said. “When they looked in the charts, they only found 10-11% of the cases had these keywords.”
Into the random forest: Kennedy is taking a different approach, using a machine-learning algorithm known as random forest (learn more about machine learning algorithms below). “It doesn’t go in looking for anything specific,” Kennedy said. “It takes a bunch of people who were diagnosed with delirium through a clinical interview, and it looks for patterns in the medical records, rather than testing hypotheses we already have.”
The average medical record contains 10,000 words. Kennedy’s algorithm picks out a tiny portion of that chart — 20-30 words — and checks to see whether they have any association with the patient’s eventual delirium diagnosis. “Then it picks another 20-30, then another 20-30, picking at random so eventually it covers all the words in the chart.”
Kennedy will repeat the procedure for all 50,000 medical records in his study. Then he plans to verify his findings in a new sample of patients admitted to the hospital.
Automated alerts: “This is down the road, but we could have a delirium risk assessment built into the electronic health record so when a person comes in to the hospital it will look through their chart and see their risk,” Kennedy said. “Then it can update that prediction during their hospital stay based on lab values or diagnoses or words that start popping up in their chart.”
2. Bringing it all together in -omics
 “UAB is very team science-oriented,” said Brittany Lasseigne, Ph.D., whose lab uses machine learning to uncover insights about disease initiation and progression. “I can go have coffee with a clinician in the Cancer Center and talk about problems they’re seeing in patients — that is something you can only do at a major medical center.”The project: Integrating Multidimensional Genomic Data to Discover Clinically Relevant Predictive Models, funded by a three-year, $747,000 R00 grant awarded in June 2019 from the National Human Genome Research Institute. Principal investigator: Brittany Lasseigne, Ph.D.
“UAB is very team science-oriented,” said Brittany Lasseigne, Ph.D., whose lab uses machine learning to uncover insights about disease initiation and progression. “I can go have coffee with a clinician in the Cancer Center and talk about problems they’re seeing in patients — that is something you can only do at a major medical center.”The project: Integrating Multidimensional Genomic Data to Discover Clinically Relevant Predictive Models, funded by a three-year, $747,000 R00 grant awarded in June 2019 from the National Human Genome Research Institute. Principal investigator: Brittany Lasseigne, Ph.D.
The problem: Genomic studies are producing a data deluge that grows exponentially with each passing year; then there are the additional avalanches generated by sequencing the transcriptome, proteome, microbiome and other -omic datasets. “Sequencers are much faster now,” Lasseigne said. “It used to take 10 days to run a batch of exomes, which only represent 3-5% of the genome. Now you can do several genomes in two days.” That’s great, but the trouble is, “you get all this data back,” Lasseigne explained. “Nobody knew what to do with it all 10 years ago and it’s worse now.
“We want to have the best picture we can have,” she continued. “Looking at only one is not the silver bullet. DNA is awesome, but outside of cancer there aren’t a lot of mutations to find. My money is on things like methylation that help us understand the tissue of origin and the broad changes we see in disease.”
The plan: “Part of the grant we’re working on is to identify other molecular patterns that are indicative of these broad changes,” Lasseigne said. “One of the reasons it’s so challenging to prevent, diagnose and treat diseases like cancer and Alzheimer’s is because everyone essentially has their own version of the disease. We’re using machine learning to see if we can group together patients where we think a common therapy will help them. We employ computers to find patterns that are meaningful for the clinic.”
Doing more with less: Another area of research in Lasseigne’s lab is cell-free sequencing. “With many types of cancer, we can’t always take a biopsy or get tissue samples of the primary tumor and we certainly can’t get samples from all the metastases,” Lasseigne said. “But they are all shedding DNA and RNA in the blood. We can take a milliliter of patient blood or urine, extract the nucleic acids and do a lot of big data analysis.”
That analysis will include a focus on the minimum requirements for diagnosis. “I don’t think we need 50 million sequencing reads to tell if a patient has cancer,” Lasseigne said. “By using smart analysis techniques and big data, we could do a profile with a million reads instead. That keeps costs down and you would need smaller samples as well.”
Lasseigne is collaborating with Eddy Yang, M.D., Ph.D., professor and vice chair for translational sciences in the Department of Radiation Oncology, senior scientist at UAB’s O’Neal Comprehensive Cancer Center and associate director for Precision Oncology in the Hugh Kaul Precision Medicine Institute. “He has patients with advanced-stage kidney cancer, and there aren’t great treatments for that,” Lasseigne said. Yang’s team collects blood samples from these patients over time, and Lasseigne looks for changes in the epigenome — “chemical modification tags on DNA,” she said. “We sequence everything there that we can and look for patterns indicating whether or not a patient is progressing or whether they are responding to a drug.”
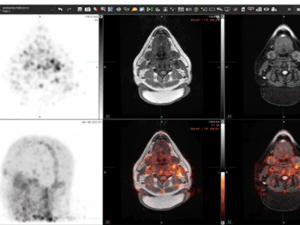
3. Whose airways will collapse?
 Surya Bhatt, M.D., is using deep learning to diagnose central airway collapse in patients with COPD. "If this is successful, it could lead to a paradigm shift" in the field, he said.The project: Deep Learning and Fluid Dynamics-Based Phenotyping of Expiratory Central Airway Collapse, funded by a $226,659 R21 grant awarded in September 2019 from the National Institute of Biomedical Imaging and Bioengineering. Principal investigator: Surya Bhatt, M.D. Investigators: Sandeep Bodduluri, Ph.D., Young-il Kim, Ph.D.
Surya Bhatt, M.D., is using deep learning to diagnose central airway collapse in patients with COPD. "If this is successful, it could lead to a paradigm shift" in the field, he said.The project: Deep Learning and Fluid Dynamics-Based Phenotyping of Expiratory Central Airway Collapse, funded by a $226,659 R21 grant awarded in September 2019 from the National Institute of Biomedical Imaging and Bioengineering. Principal investigator: Surya Bhatt, M.D. Investigators: Sandeep Bodduluri, Ph.D., Young-il Kim, Ph.D.
The problem: More than 16 million Americans have been diagnosed with chronic obstructive pulmonary disease (COPD), according to the Centers for Disease Control and Prevention. Millions more suffer from the disease, which is characterized by blocked airflow and breathing-related problems. Alabama has some of the highest rates of COPD in the United States.
For most people with COPD, the main site of blockage is in the small airways of the lungs. But for some 5% of patients, breathing problems may be caused by collapse of the large airways when they exhale. The distinction is crucial, because bronchodilators, which are the first-line treatment for COPD, are effective in expanding the small airways but they do not help when the problem is collapsed large airways. “Unfortunately, the symptoms are almost exactly the same,” Bhatt said. “It’s very difficult to identify these patients in clinical practice. We estimate that there are 3.5 to 4 million people in the United States with this condition. It is significantly underappreciated.”
Currently, when physicians suspect large airway collapse they can investigate with bronchoscopy or with two CT scans — one as they hold their breath at full inhalation and another after they have completely exhaled. The first method is invasive and the second involves a substantial amount of radiation exposure to the patient.
Seeking a third way with deep learning: With his R21 exploratory grant, Bhatt is exploring a third way. “We want to know, can we identify these patients with just a single CT scan, by training a machine to look at the geometry of airways and identify the probability that their airways will collapse when they breathe out,” he said.
The plan: Bhatt has a large database of CT images that includes individuals with large airway collapse. “We’re going to feed the raw images into the computer and use deep learning to identify features that humans haven’t been able to pick up: subtle changes in branching patterns or geometry of the airways or differences in the wall,” Bhatt said. “That’s the hope.”
The work is done on Cheaha and also with an Nvidia computer in the lab, Bhatt said. Much of the programming is done by his trainee, Sandeep Bodduluri, Ph.D., an instructor in the division.
The endgame: “If this is successful, it could lead to a paradigm shift” in the field, Bhatt said. “Right now we don’t even identify most of these patients. If you have a high clinical suspicion with unexplained symptoms, this algorithm could give a probability score for someone to have this problem and then the physician could investigate further.”
Learn the basics of machine learning with hands-on videos from UAB’s Data Science Club

Machine-learning helps predict which cancer patients are most likely to enter the fog
DREAM Challenge to automate assessment of radiographic damage from rheumatoid arthritis

 Machine learning has a bewildering array of approaches to choose from. But “ultimately it all comes down to trees or regression — making a tree out of the data or fitting a line to the data,” explains Brittany Lasseigne, Ph.D. Here are some of the most popular algorithms used in the field and in UAB labs.
Machine learning has a bewildering array of approaches to choose from. But “ultimately it all comes down to trees or regression — making a tree out of the data or fitting a line to the data,” explains Brittany Lasseigne, Ph.D. Here are some of the most popular algorithms used in the field and in UAB labs.